Technological
Innovation
We interrogate the metabolome with unprecedented depth, confidence, and quantitative accuracy by exploiting an armamentarium of advanced analytical approaches.
Research in the Patti Lab
Our work is highly interdisciplinary, spanning three complementary areas: cancer biology, technology development, and computational biology. Team members have diverse scientific backgrounds and interests but have synergistic goals that promote collaborative activities within the group. The result is a vibrant training environment where investigators from different disciplines continuously learn from one another. Collectively, the team’s breadth of expertise provides unique scientific perspective and makes us particularly well suited to address some of the grandest challenges in biology and medicine.

Technology Development
The Patti lab pioneers new technologies and experimental strategies that empower us to pursue previously intractable biological questions. An overarching goal is to extend the scale over which metabolism can be studied. At one end of the spectrum, we seek to establish mass spectrometry-based profiling approaches that are amenable to large, diverse, and longitudinal cohorts of tens of thousands or hundreds of thousands of individuals. This includes performing untargeted metabolomics, non-targeted analysis of chemical exposures, and discovery proteomics at the population level in collaboration with partners such as the UK Biobank. We believe that such large-scale experiments are essential for precision medicine to identify subgroups within the population for whom strategies to prevent, diagnose, and treat disease can be uniquely tailored.
At the opposite end of the spectrum, we want to measure metabolites and proteins from single cells by using mass spectrometry-based technologies such as in situ imaging. Perhaps in no other area is single-cell metabolomics poised to have as transformative an impact as in cancer. Tumors contain diverse populations of cancer cells, vascular cells, fibroblasts, immune cells, and cancer stem cells. Yet, due to limitations in conventional methods, almost everything we know about cancer metabolism is based on analyzing metabolites and proteins from tumors in bulk or individual cell lines in culture. Our goal is to map metabolic interactions in cancer at the single-cell level. We seek not only to understand how adjacent cells within a tissue are biochemically coupled but also how cells in anatomically distant organs engage in molecular crosstalk. An integral part of our effort to uncover the complex metabolic relationships that cells entertain with each other is developing novel strategies to introduce stable isotope tracers into model organisms and human patients. In addition to mice, we rely heavily on zebrafish and have established innovative approaches for isotope labeling, mass spectrometry-based tissue imaging, and high-throughput screening of environmental chemicals that promote tumorigenesis in adult animals.

QTOF and Orbitrap Mass Spectrometers
Our quadrupole time-of-flight and Orbitrap instruments provide high-resolution data that can be used to profile water-soluble metabolites, lipids, and chemical exposures at the comprehensive scale. We also use Orbitrap instruments for discovery proteomics.

Orbitrap Astral Mass Spectrometers
These instruments combine a quadrupole, Orbitrap, and Astral mass analyzer to achieve faster and more sensitive measurements. They are particularly well suited for discovery proteomics profiling.

Ion Mobility Spectrometry
Compound identification is enhanced by matching collision cross sections to authentic standards. Our technologies include trapped ion mobility spectrometry, traveling wave ion mobility spectrometry, and drift tube ion mobility spectrometry.

Seer Proteograph
The Proteograph provides an automated workflow to prepare proteomics samples efficiently, thereby facilitating large-scale studies. Combining Seer’s nanoparticle technology with liquid chromatography/mass spectrometry extends the depth of proteome coverage when profiling biofluids.
Mass Spectrometry Imaging
In situ metabolomics localizes compounds to specific anatomical regions or cells within a tissue. We perform mass spectrometry imaging by using standard MALDI and DESI instruments from the factory as well as by retrofitting mass spectrometers with technologies such as MassTech and NIMS.

Instruments for Targeted Analysis
Historically, targeted analysis has been limited to a relatively small number of analytes . New triple quadrupole and Stellar instruments are so fast, however, that large panels of small molecules and proteins can be assessed in a single analysis. These instruments are expanding our application space of targeted workflows, facilitating validation and absolute quantitation experiments

High-Resolution Fluorescence Microscopy
Our Leica Thunder imager uses a new Instant Computational Clearing method to generate high-resolution and high-contrast images for 3D visualization of biological processes in cells and tissues.
In-House Zebrafish Facility
We use a zebrafish Tecniplast system to house genetic models of cancer and enable in vivo isotope tracing.

Respirometry
Seahorse extracellular flux analyzers provide us with insight into oxygen consumption and other discrete changes in bioenergetics in real time.
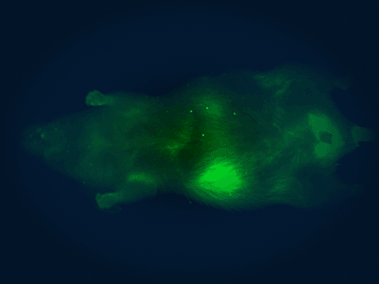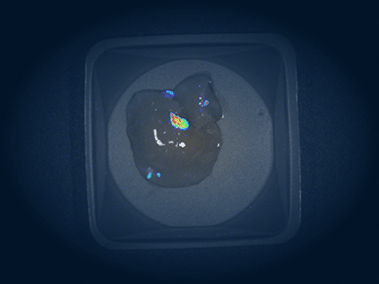
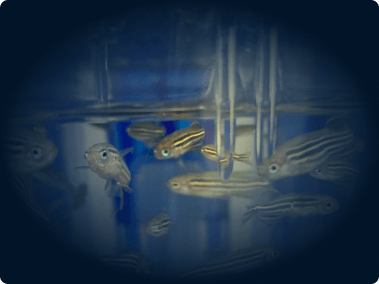
In vivo imaging system
The Pearl Triology imaging system enables bioluminescence detection and dual-channel near-infrared fluorescent detection, which we use for monitoring the location, progression, and therapeutic response of tumors in mice.
Vendors
Our workflows use in-house informatics solutions, home-built equipment, and instrumentation from vendors including:
Software
Mass spectrometry data produced from the analysis of biological samples are highly complex. It is not practical to perform a systems-level analysis of the results by manual evaluation. The Patti Laboratory develops software solutions to automate data processing and enhance interpretation.